SWNTGenerator¶
-
class
sknano.generators.SWNTGenerator(*args, from_scaling_matrix=False, **kwargs)[source] [edit on github][source]¶ Bases:
sknano.generators.NanotubeBundleGeneratorBase,sknano.generators.SWNTGeneratorBase,sknano.core.structures.SWNTClass for generating
SWNTstructures.Parameters: - *Ch –
Either a 2-tuple of integers (i.e., *Ch = [(n, m)]) or 2 integers (i.e., *Ch = [n, m]) specifying the chiral indices of the nanotube chiral vector \(\mathbf{C}_h = n\mathbf{a}_1 + m\mathbf{a}_2 = (n, m)\).
- ny, nz (nx,) – Number of repeat unit cells in the \(x, y, z\) dimensions.
- basis ({
list}, optional) –List of
strs of element symbols or atomic number of the two atom basis (default: [‘C’, ‘C’])New in version 0.3.10.
- element2 (element1,) –
Element symbol or atomic number of basis
Atom1 and 2Deprecated since version 0.3.10: Use
basisinstead - bond (float, optional) – \(\mathrm{a}_{\mathrm{CC}} =\) distance between nearest neighbor atoms. Must be in units of Angstroms.
- Lz (float, optional) –
Length of nanotube in units of Angstroms. Overrides the
nzvalue.New in version 0.2.5.
Changed in version 0.4.0: Changed units from nanometers to Angstroms
- tube_length (float, optional) –
Length of nanotube in units of Angstroms. Overrides the
nzvalue.Deprecated since version 0.2.5: Use
Lzinstead - fix_Lz (bool, optional) –
Generate the nanotube with length as close to the specified \(L_z\) as possible. If
True, then non integer \(n_z\) cells are permitted.New in version 0.2.6.
- vdw_radius (float, optional) –
van der Waals radius of nanotube atoms
New in version 0.2.5.
- bundle_packing ({'hcp', 'ccp'}, optional) –
Packing arrangement of nanotubes bundles. If
bundle_packingisNone, then it will be determined by thebundle_geometryparameter ifbundle_geometryis notNone. If bothbundle_packingandbundle_geometryareNone, thenbundle_packingdefaults tohcp.New in version 0.2.5.
- bundle_geometry ({'triangle', 'hexagon', 'square', 'rectangle'}, optional) –
Force a specific geometry on the nanotube bundle boundaries.
New in version 0.2.5.
- autogen (bool, optional) – if
True, automatically generate structure data. - verbose (bool, optional) – if
True, show verbose output
Examples
Using the
SWNTGeneratorclass, you can generate structure data for single-walled nanotubes (SWNTs) or nanotube bundles with either cubic close packed (ccp) or hexagonal close packed (hcp) arrangement of nanotubes. The bundle packing arrangement is set using thebundle_packingparameter.You can also enforce a specific
bundle geometrywhich will try and build the nanotube bundle such that it “fits inside” the boundaries of a specified geometric shape. This allows you to generate hcp bundles that are trianglar, hexagonal, or rectangular in shape, as some of the examples below illustrate.First, load the
SWNTGeneratorclass.>>> from sknano.generators import SWNTGenerator
Now let’s generate a \(\mathbf{C}_{\mathrm{h}} = (10, 5)\)
SWNTunit cell.>>> swnt = SWNTGenerator((10, 5)) >>> swnt.save(fname='10,5_unit_cell.xyz') >>> # note that there are two other alternative, but equivalent >>> # means of passing arguments to SWNTGenerator constructor: >>> # SWNTGenerator(10, 5) and SWNTGenerator(n=10, m=5)
Here’s a nice ray traced rendering of the generated \((10, 5)\)
SWNTunit cell.
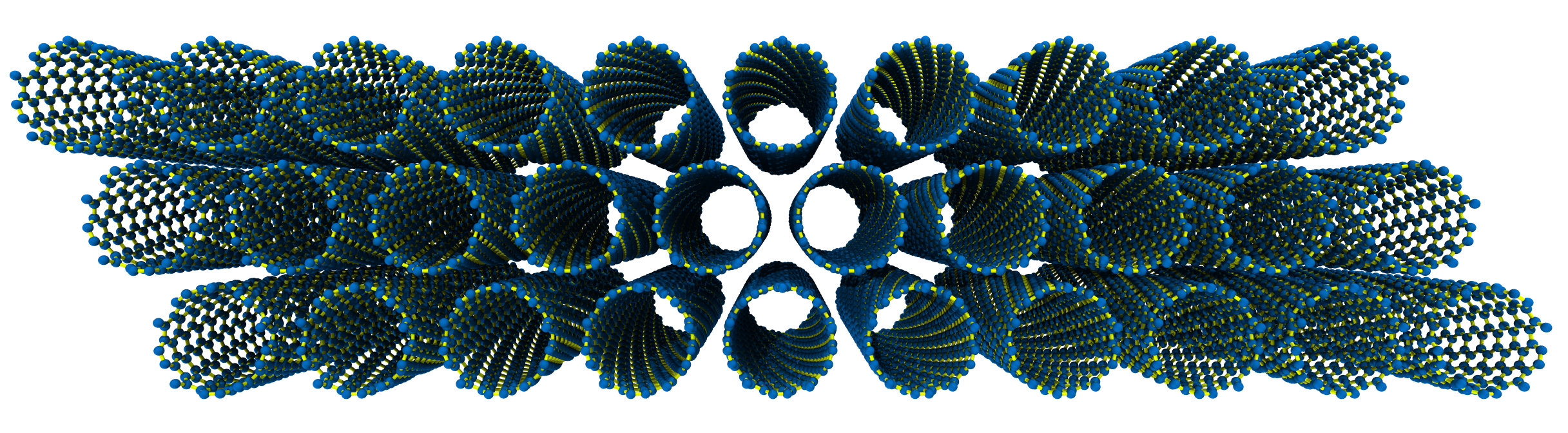
Next, let’s generate an hcp bundle of \(C_{\mathrm{h}} = (10, 5)\) SWCNTs and cell count \(n_x=10, n_y=3, n_z=5\).
>>> SWCNTbundle = SWNTGenerator(n=10, m=5, nx=10, ny=3, nz=5) >>> SWCNTbundle.save()
The rendered structure looks like:
Now let’s generate a nice hexagon bundle, 3 tubes wide, with \(C_{\mathrm{h}} = (6, 5)\).
>>> SWCNTbundle = SWNTGenerator(n=6, m=5, nz=5, bundle_geometry='hexagon') >>> SWCNTbundle.save()
which looks like:

Remember, all of the
savemethods allow you to rotate the structure data before saving:>>> SWCNTbundle.save(fname='0605_hcp_7tube_hexagon_rot-30deg.xyz', ... rot_axis='z', rotation_angle=30)

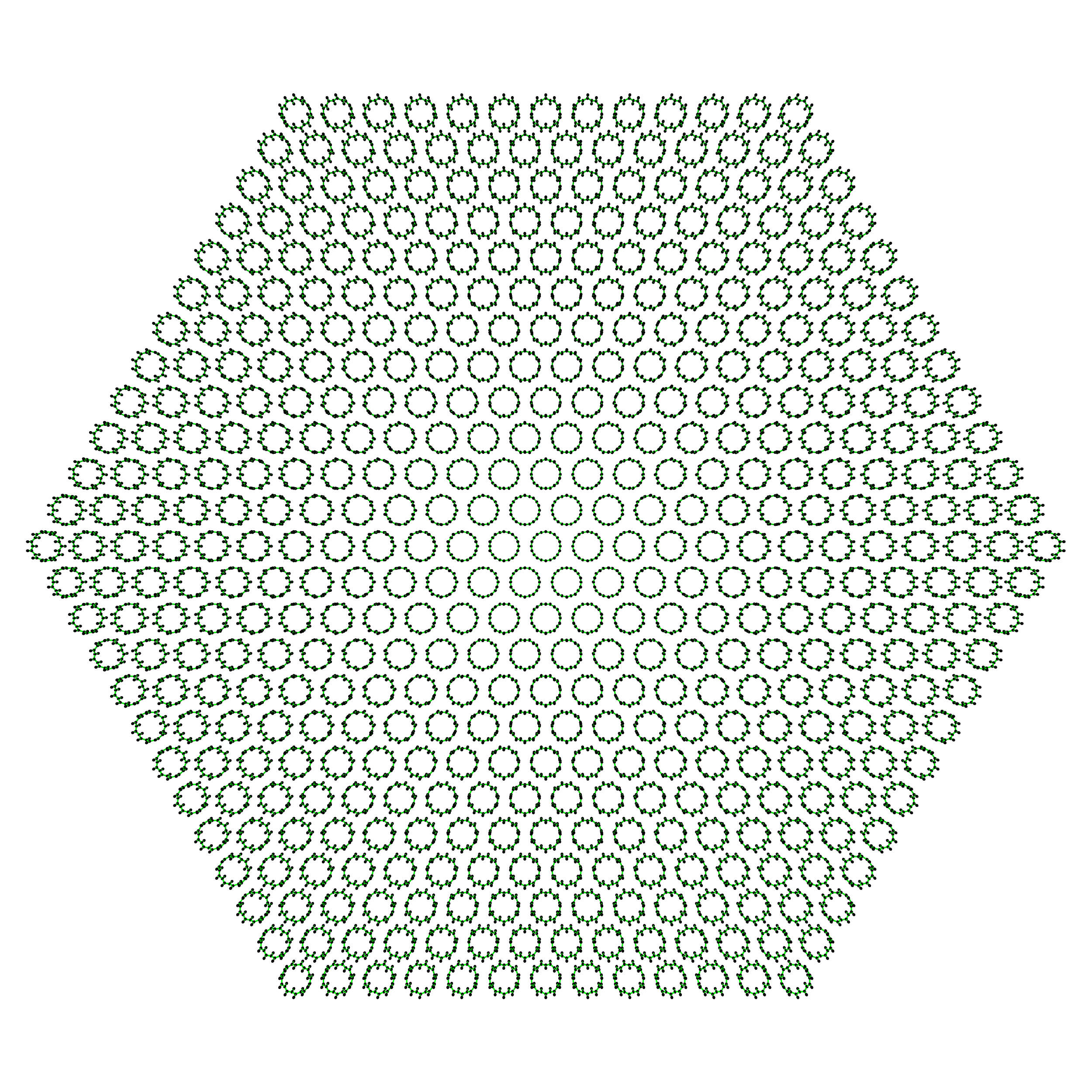
Now, just because we can, let’s make a big ass hexagon bundle with \(C_{\mathrm{h}} = (10, 0)\).
>>> big_hexagonal_bundle = SWNTGenerator(10, 0, nx=25, ny=25, nz=1, ... bundle_geometry='hexagon') >>> big_hexagonal_bundle.save()
Take a look at the 469 \((10, 0)\) unit cells in this big ass bundle!
Lastly, here’s a look at a bundle generated with cubic close packed bundle arrangement:
>>> SWCNTbundle = SWNTGenerator(10, 10, nx=3, ny=3, nz=5, ... bundle_packing='ccp') >>> SWCNTbundle.save()
The rendered
ccpstructure looks like: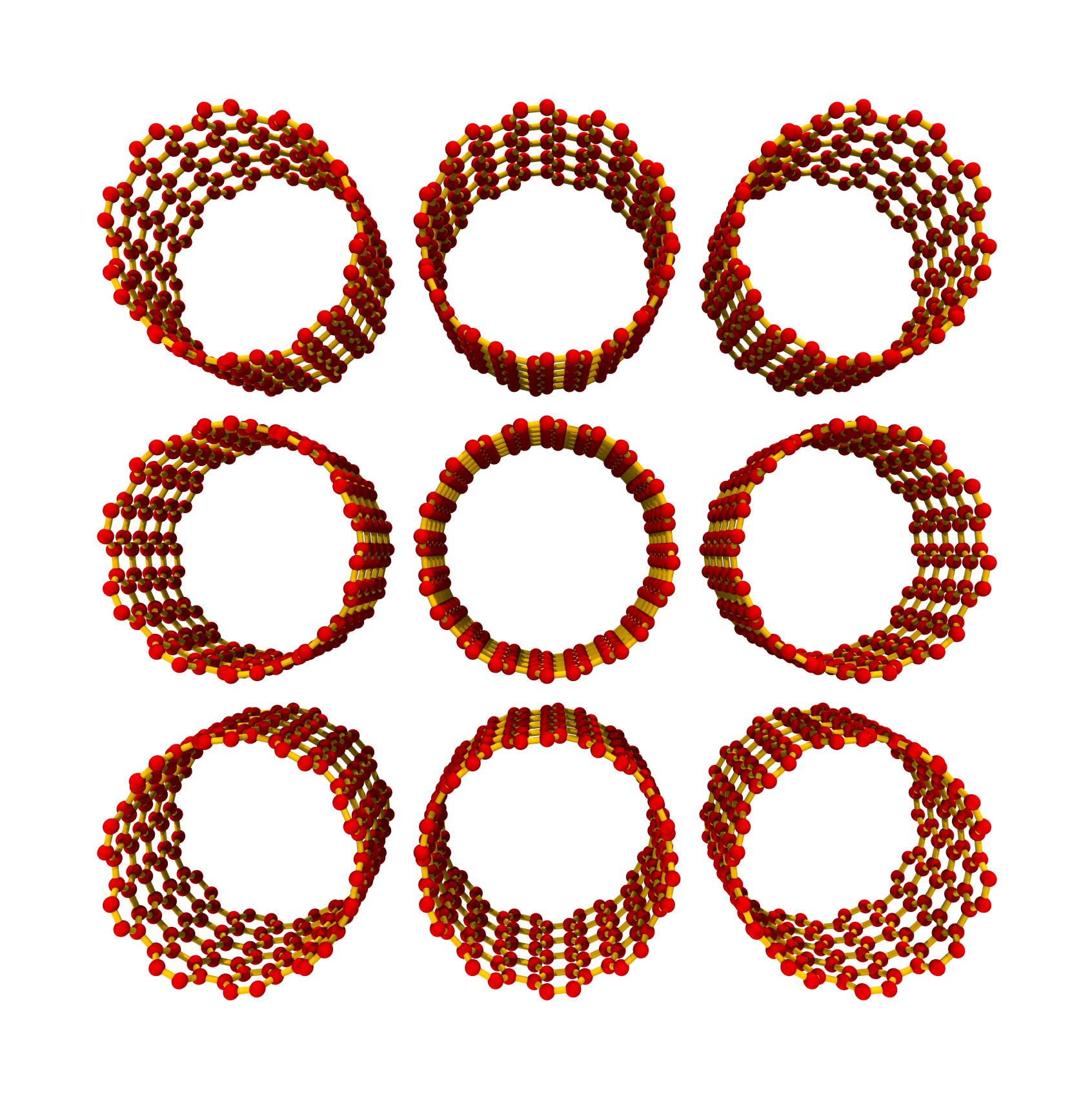
Attributes
ChSWNT circumference \(|\mathbf{C}_h|\) in Å Ch_vecSWNT chiral vector. LxAxis-aligned length along the x-axis in Angstroms.LyAxis-aligned length along the y-axis in Angstroms.LzSWNT length \(L_z = L_{\mathrm{tube}}\) in Angstroms. M\(M = np - nq\) NNumber of graphene hexagons in nanotube unit cell. NatomsN atoms. Natoms_listlistofNatomsper nanotube in bundle.Natoms_per_bundleAlias for Natoms.Natoms_per_tubeAlias for Natoms_list.Natoms_per_unit_cellNumber of atoms in nanotube unit cell. NtubesNumber of nanotubes in bundle. RSymmetry vector \(\mathbf{R} = (p, q)\). TLength of nanotube unit cell \(|\mathbf{T}|\) in Å. TvecSWNTtranslation vector.atomsStructure Atoms.basisNanoStructureBasebasis objects.bundle_densityCompute the nanotube bundle density. bundle_geometryBundle geometry. bundle_massAn alias for mass.bundle_packingBundle packing. chiral_angleChiral angle \(\theta_c\) in degrees. chiral_typeSWNTchiral type.crystal_cellStructure CrystalCell.d\(d=\gcd{(n, m)}\) dR\(d_R=\gcd{(2n + m, 2m + n)}\) dtNanotube diameter \(d_t = \frac{|\mathbf{C}_h|}{\pi}\) in Å. electronic_typeSWNT electronic type. element1Basis element 1 element2Basis element 2 fix_Lzboolindicating whetherSWNTMixin.Lzis fixed or calculated.fmtstrFormat string. latticeStructure Crystal3DLattice.lattice_shiftLattice displacement vector. linear_mass_densityLinear mass density of nanotube in g/Å. mChiral index \(m\). massTotal mass of atoms. nChiral index \(n\). nxNumber of nanotubes along the \(x\)-axis. nyNumber of nanotubes along the \(y\)-axis. nzNumber of nanotube unit cells along the \(z\)-axis. rtNanotube radius \(r_t = \frac{|\mathbf{C}_h|}{2\pi}\) in Å. scaling_matrixCrystalCell.scaling_matrix.structureAn alias to self.t1\(t_{1} = \frac{2m + n}{d_{R}}\) t2\(t_2 = -\frac{2n + m}{d_R}\) tube_lengthAlias for SWNT.Lztube_massAn alias for mass.unit_cellStructure UnitCell.unit_cell_massUnit cell mass in atomic mass units. unit_cell_symmetry_paramsTuple of SWNTunit cell symmetry parameters.vdw_distanceVan der Waals distance. vdw_radiusVan der Waals radius Methods
clear()Clear list of StructureMixin.atoms.finalize()Finalize structure data by clipping region bounds if fix_LzisTrue.generate([finalize])Generate structure data. generate_bundle_coords()Generate coordinates of bundle tubes. generate_fname([n, m, nx, ny, nz, fix_Lz, ...])Generate filename string. init_bundle_parameters()Initialize bundle attributes. make_supercell(scaling_matrix[, wrap_coords])Make supercell. rotate(**kwargs)Rotate crystal cell lattice, basis, and unit cell. save([fname, outpath, structure_format, ...])Save structure data. todict()Return dictof constructor parameters.transform_lattice(scaling_matrix[, ...])Transform structure lattice. translate(t[, fix_anchor_points])Translate crystal cell lattice, basis, and unit cell. write(*args, **kwargs)Write structure data to file. write_data(**kwargs)Write LAMMPS data file. write_dump(**kwargs)Write LAMMPS dump file. write_pdb(**kwargs)Write pdb file. write_xyz(**kwargs)Write xyz file. Methods Summary
finalize()Finalize structure data by clipping region bounds if fix_LzisTrue.generate_fname([n, m, nx, ny, nz, fix_Lz, ...])Generate filename string. save([fname, outpath, structure_format, ...])Save structure data. Methods Documentation
-
finalize()[source] [edit on github][source]¶ Finalize structure data by clipping region bounds if
fix_LzisTrue.
-
classmethod
generate_fname(n=None, m=None, nx=None, ny=None, nz=None, fix_Lz=False, Ntubes=None, bundle_geometry=None, bundle_packing=None, **kwargs)[source] [edit on github][source]¶ Generate filename string.
-
save(fname=None, outpath=None, structure_format=None, center_centroid=True, **kwargs)[source] [edit on github][source]¶ Save structure data.
See
savemethod for documentation.
- *Ch –
