sknano.generators.SWNTBundleGenerator¶
-
class
sknano.generators.SWNTBundleGenerator(autogen=True, **kwargs)[source][source]¶ Class for generating nanotube bundles.
New in version 0.2.4.
Parameters: Either a 2-tuple of ints or 2 integers giving the chiral indices of the nanotube chiral vector \(\mathbf{C}_h = n\mathbf{a}_1 + m\mathbf{a}_2 = (n, m)\).
nx, ny, nz : int, optional
Number of repeat unit cells in the \(x, y, z\) dimensions.
basis : {
list}, optionalList of
strs of element symbols or atomic number of the two atom basis (default: [‘C’, ‘C’])New in version 0.3.10.
element1, element2 : {str, int}, optional
Element symbol or atomic number of basis
Atom1 and 2Deprecated since version 0.3.10: Use
basisinsteadbond : float, optional
\(\mathrm{a}_{\mathrm{CC}} =\) distance between nearest neighbor atoms. Must be in units of Angstroms.
vdw_radius : float, optional
van der Waals radius of nanotube atoms
New in version 0.2.5.
bundle_packing : {‘hcp’, ‘ccp’}, optional
Packing arrangement of nanotubes bundles. If
bundle_packingis None, then it will be determined by thebundle_geometryparameter ifbundle_geometryis not None. If bothbundle_packingandbundle_geometryare None, thenbundle_packingdefaults to hcp.New in version 0.2.5.
bundle_geometry : {‘triangle’, ‘hexagon’, ‘square’, ‘rectangle’}, optional
Force a specific geometry on the nanotube bundle boundaries.
New in version 0.2.5.
Lz : float, optional
length of bundle in \(z\) dimension in nanometers. Overrides
nzvalue.New in version 0.2.5.
fix_Lz : bool, optional
Generate the nanotube with length as close to the specified \(L_z\) as possible. If True, then non integer \(n_z\) cells are permitted.
New in version 0.2.6.
autogen : bool, optional
if True, automatically generate structure data.
verbose : bool, optional
if True, show verbose output
Examples
Using the
SWNTBundleGeneratorclass, you can generate structure data for nanotube bundles with either cubic close packed (ccp) or hexagonal close packed (hcp) arrangement of nanotubes. The bundle packing arrangement is set using thebundle_packingparameter.You can also enforce a specific bundle geometry which will try and build the nanotube bundle such that it “fits inside” the boundaries of a specified geometric shape. This allows you to generate hcp bundles that are trianglar, hexagonal, or rectangular in shape, as some of the examples below illustrate.
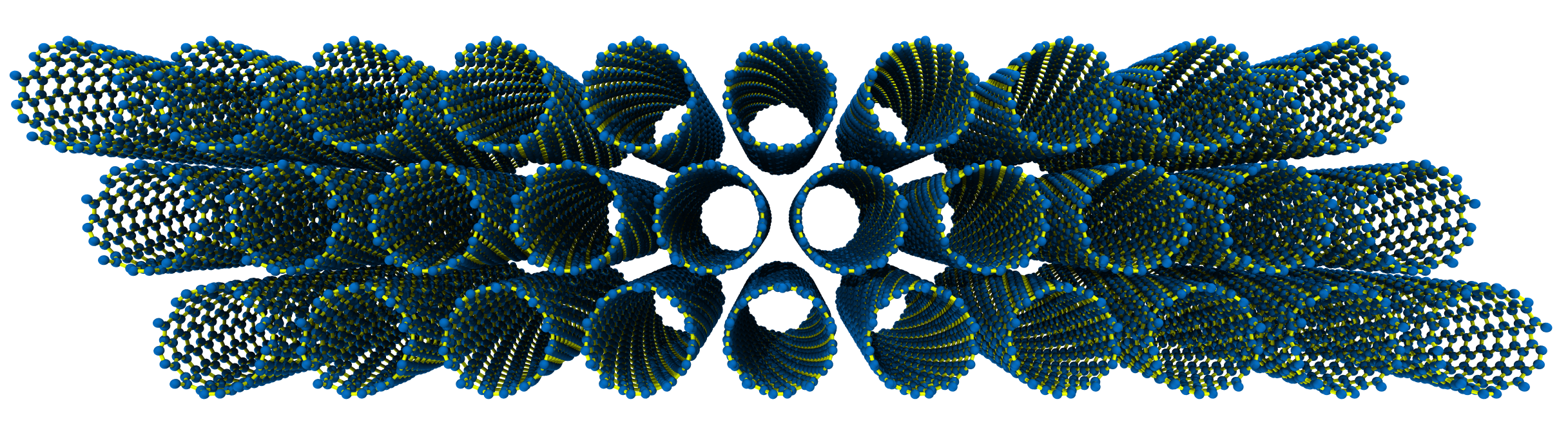
To start, let’s generate an hcp bundle of \(C_{\mathrm{h}} = (10, 5)\) SWCNTs and cell count \(n_x=10, n_y=3, n_z=5\).
>>> from sknano.generators import SWNTBundleGenerator >>> SWCNTbundle = SWNTBundleGenerator(n=10, m=5, nx=10, ny=3, nz=5) >>> SWCNTbundle.save()
The rendered structure looks like:
Now let’s generate a nice hexagon bundle, 3 tubes wide, with \(C_{\mathrm{h}} = (6, 5)\).
>>> SWCNTbundle = SWNTBundleGenerator(n=6, m=5, nz=5, ... bundle_geometry='hexagon') >>> SWCNTbundle.save()
which looks like:

Remember, all of the
savemethods allow you to rotate the structure data before saving:>>> SWCNTbundle.save(fname='0605_hcp_7tube_hexagon_rot-30deg.xyz', ... rot_axis='z', rotation_angle=30)

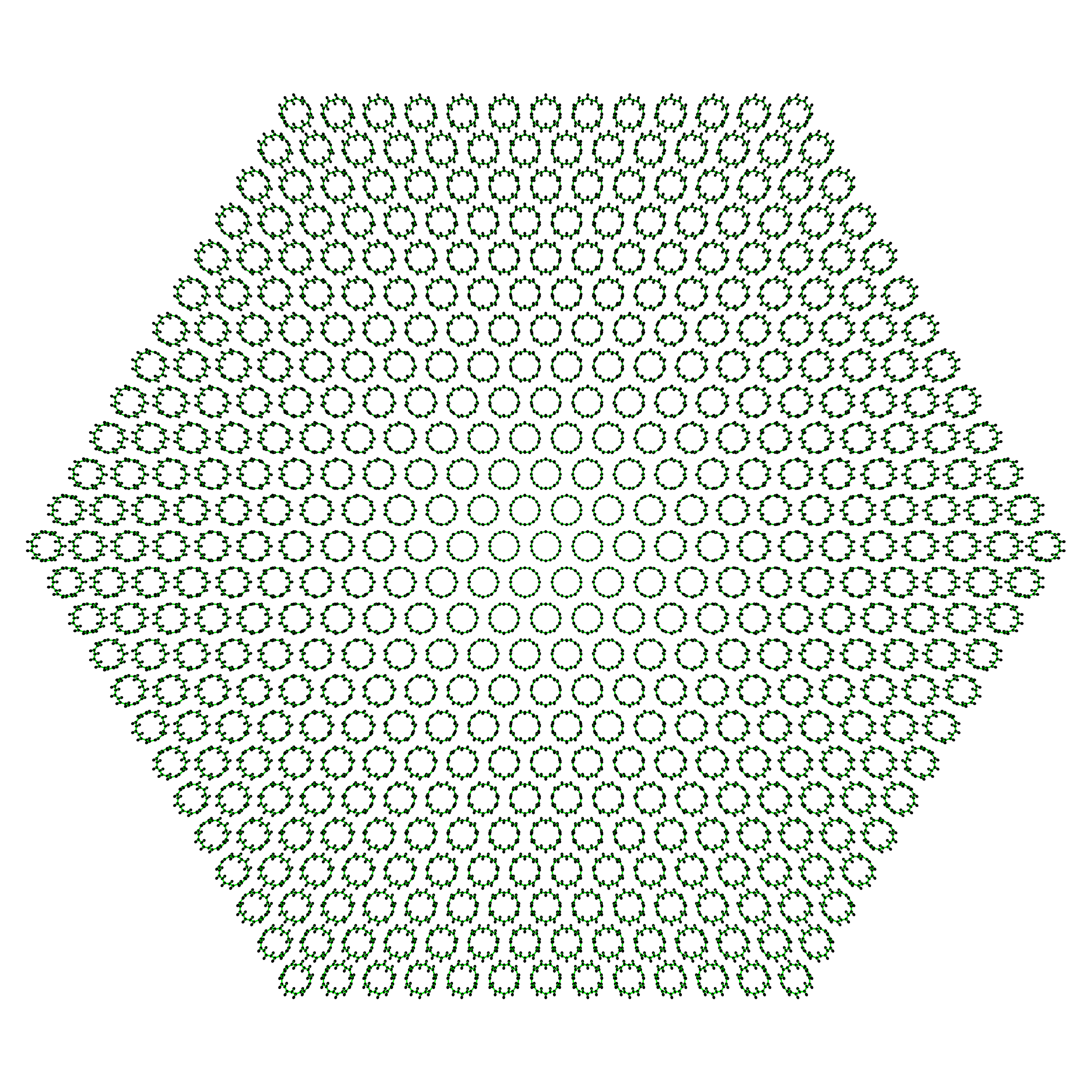
Now, just because we can, let’s make a big ass hexagon bundle with \(C_{\mathrm{h}} = (10, 0)\).
>>> big_hexagonal_bundle = SWNTBundleGenerator(10, 0, nx=25, ny=25, nz=1, ... bundle_geometry='hexagon') >>> big_hexagonal_bundle.save()
Take a look at the 469 \((10, 0)\) unit cells in this big ass bundle!
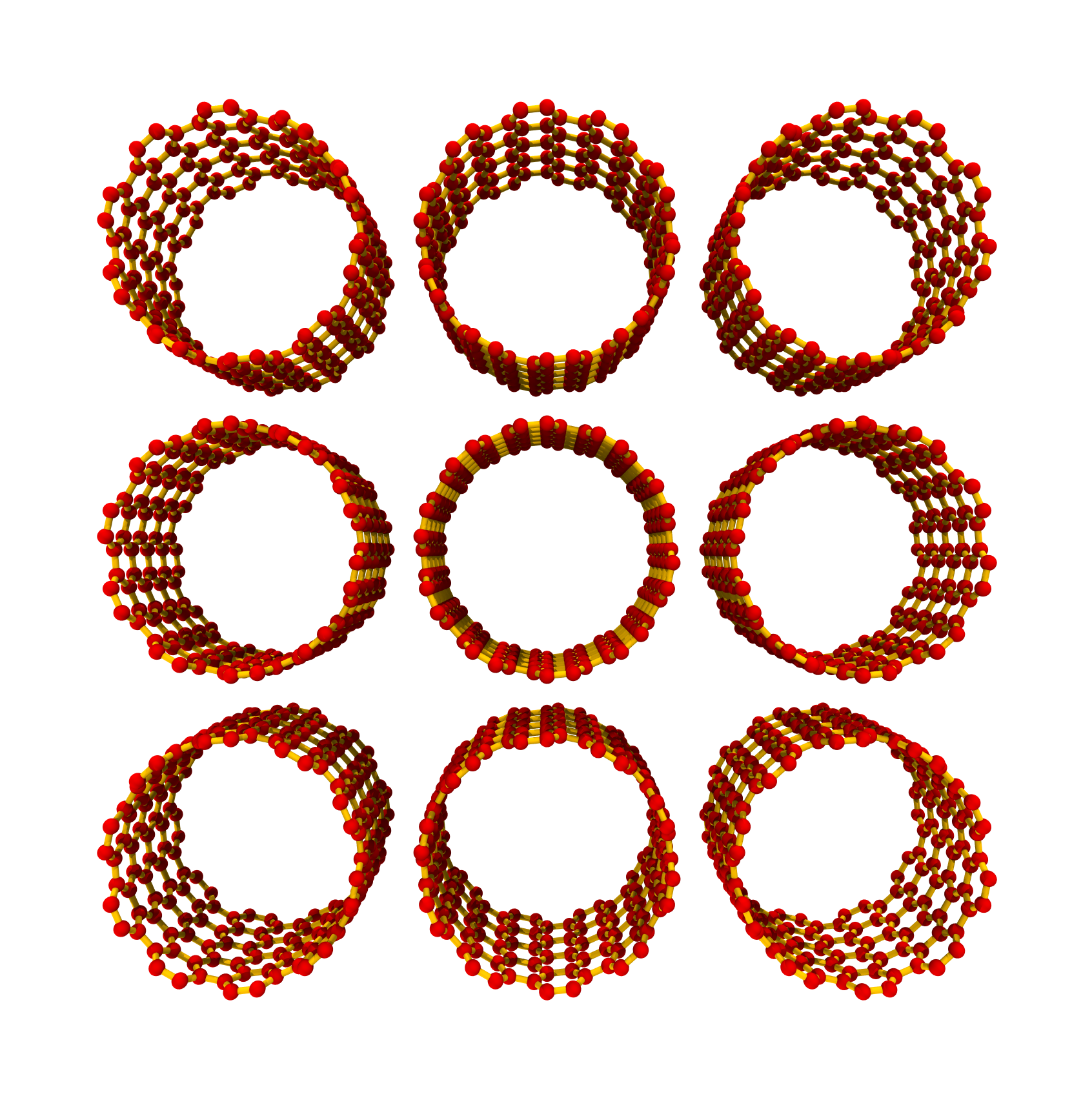
Lastly, here’s a look at a bundle generated with cubic close packed bundle arrangement:
>>> SWCNTbundle = SWNTBundleGenerator(10, 10, nx=3, ny=3, nz=5, ... bundle_packing='ccp') >>> SWCNTbundle.save()
The rendered ccp structure looks like:
Attributes
ChSWNT circumference \(|\mathbf{C}_h|\) in Å Ch_vecSWNT chiral vector. LxLyLzSWNT length \(L_z = L_{\mathrm{tube}}\) in nanometers. M\(M = np - nq\) NNumber of graphene hexagons in nanotube unit cell. NatomsNumber of atoms in nanotube bundle. Natoms_listNatoms_per_bundleNatoms_per_tubeAlias for Natoms_list.Natoms_per_unit_cellNumber of atoms in nanotube unit cell. NtubesRSymmetry vector \(\mathbf{R} = (p, q)\). TLength of nanotube unit cell \(|\mathbf{T}|\) in Å. TvecSWNT translation vector. atomsStructure StructureAtoms.basisNanoStructureBasebasis atoms.bundle_densitybundle_geometrybundle_massbundle_packingchiral_angleChiral angle \(\theta_c\) in degrees. chiral_typeSWNT chiral type. crystal_cellStructure CrystalCell.d\(d=\gcd{(n, m)}\) dR\(d_R=\gcd{(2n + m, 2m + n)}\) dtNanotube diameter \(d_t = \frac{|\mathbf{C}_h|}{\pi}\) in Å. electronic_typeSWNT electronic type. element1Basis element 1 element2Basis element 2 fix_LzfmtstrFormat string. latticeStructure Crystal3DLattice.linear_mass_densityLinear mass density of nanotube in g/nm. mChiral index \(m\). nChiral index \(n\). nxNumber of nanotubes along the \(x\)-axis. nyNumber of nanotubes along the \(y\)-axis. nzNumber of nanotube unit cells along the \(z\)-axis. rtNanotube radius \(r_t = \frac{|\mathbf{C}_h|}{2\pi}\) in Å. scaling_matrixCrystalCell.scaling_matrix.structurePointer to self. structure_dataAlias for BaseStructureMixin.structure.t1\(t_{1} = \frac{2m + n}{d_{R}}\) t2\(t_2 = -\frac{2n + m}{d_R}\) tube_lengthAlias for SWNT.Lztube_massSWNT mass in grams. unit_cellStructure UnitCell.unit_cell_massUnit cell mass in atomic mass units. unit_cell_symmetry_paramsTuple of SWNT unit cell symmetry parameters. vdw_distancevan der Waals distance. vdw_radiusvan der Waals radius Methods
clear()Clear list of BaseStructureMixin.atoms.generate([generate_bundle, ...])Generate structure data. generate_bundle()generate_bundle_coords()Generate coordinates of bundle tubes. generate_bundle_from_bundle_coords()generate_fname([n, m, nx, ny, nz, fix_Lz, ...])generate_unit_cell()Generate the nanotube unit cell. make_supercell(scaling_matrix[, wrap_coords])Make supercell. read_data(*args, **kwargs)read_dump(*args, **kwargs)read_xyz(*args, **kwargs)rotate(**kwargs)Rotate crystal cell lattice, basis, and unit cell. save([fname, outpath, structure_format, ...])Save structure data. todict()transform_lattice(scaling_matrix[, ...])translate(t[, fix_anchor_points])Translate crystal cell basis. write_data(**kwargs)write_dump(**kwargs)write_xyz(**kwargs)
